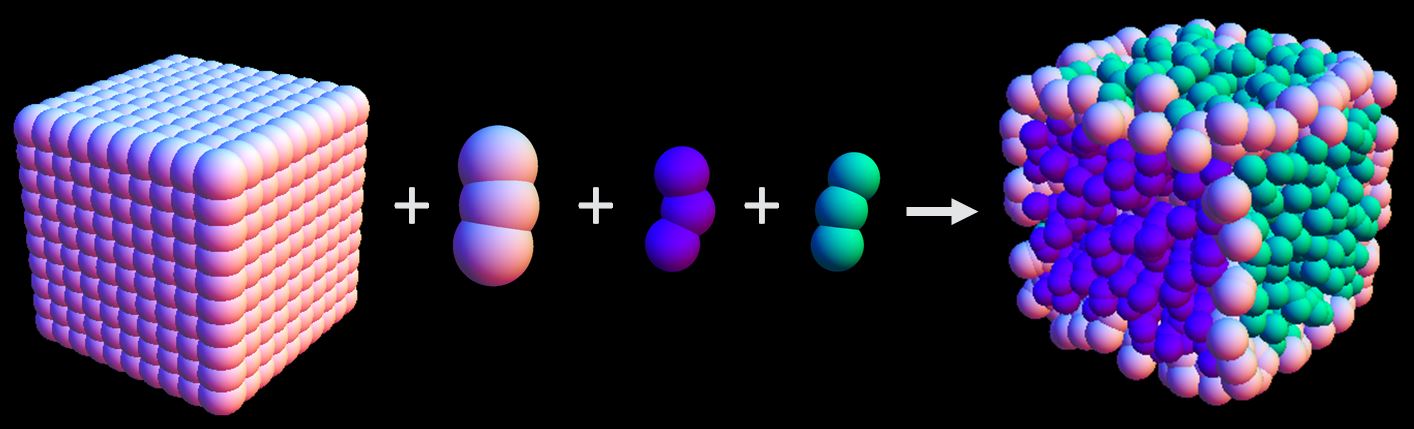Design + Assembly
In order to simplify design and assembly, we first build nanocubes that can be programmed to bind face-to-face. Like LEGOs, large complex structures can be built by connecting smaller assemblages of blocks together. In fact, DNP123 cubes are even easier to build with than LEGOs, because they self-assemble. Unlike lithography, which requires expensive clean rooms and heavy equipment, no complex machinery is needed to connect the cubes.
To program assembly, each nanocube face is patched with a unique sequence of single stranded DNA. Target structures can be designed by selecting which faces from different cubes should bind together. When placed in solution, the lock-and-key binding of complementary DNA ligands ensures that only the target structures will form.
To program assembly, each nanocube face is patched with a unique sequence of single stranded DNA. Target structures can be designed by selecting which faces from different cubes should bind together. When placed in solution, the lock-and-key binding of complementary DNA ligands ensures that only the target structures will form.
Step 1: Make Blocks
Nanocubes are synthesized and then their faces are patched. Each face of a nanocube can be patched by first drying the cubes on a flat surface and then applying a stamp coated in thiolated DNA to the top face. By repeatedly stamping six times with unique DNA sequences, we coat each face with a one-of-a-kind address that can be targeted for binding. Yield can be improved dramatically by appropriately selecting the surface chemistry of the flat surface.
Step 2: Program Assembly
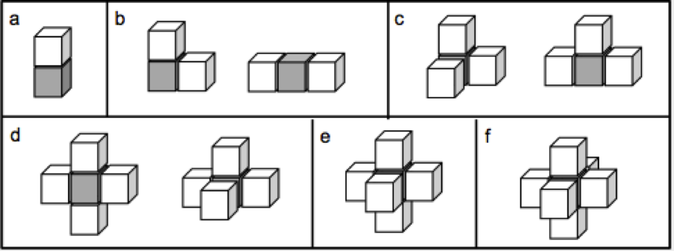
By patching each face with a unique strand of DNA, we encode unambiguous instructions for assembly. When combined in solution, only cube faces patched with complimentary sequences will bind face-to-face. This allows one to program how the cubes will assemble into a larger 3D structure. A complete set of all possible binding structures is possible, so one can design any conceivable shape.
Step 3: Build the device
Once cubes are synthesized and programmed to form the desired structure, assembly is simply a matter of mixing solutions of the cubes together. The cubes will handle the rest of the work as they self-assemble into the desired product.
Home |
About |
Services |
Menu |
Contact |
Copyright © 2018